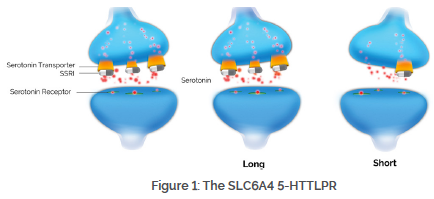
What is SLC6A4?
The serotonin transporter, encoded by the SLC6A4 gene, is responsible for serotonin reuptake into the presynaptic neuron. While many antidepressants have some serotonin reuptake blocking activity, it is thought to be the major mechanism of action of the selective serotonin reuptake inhibitors (SSRIs).
Another variant, rs25531, is a single A>G substitution near the promoter region of SLC6A4. An initial study found that this allele stratified the long form of the gene into two groups: high transcriptional activity (LA) and transcriptional activity comparable to the short allele (LG).1 However, subsequent attempts to replicate this finding have failed.2,3 Nevertheless, many studies have genotyped the rs25531 SNP in conjunction with the 5-HTTLPR to measure their impact on SSRI efficacy and adverse events, producing mixed results.
Is there a connection between SLC6A4 genotype and antidepressant efficacy?
Multiple meta-analyses have reviewed the impact of the 5-HTTLPR on antidepressant efficacy. Most recently, Karlovic et al. reviewed results from 35 studies and 3 meta-analyses (n=8,424).4 The results were divided by medication class and ethnicity. Among Caucasians taking SSRIs, 14 of 17 studies showed an impact of the 5-HTTLPR on treatment outcome. Seven of those 14 studies found that carriers of the S allele (L/S or S/S) had a poorer outcome when taking SSRIs, while the other 7 found the S/S genotype to have a poorer outcome. In contrast, analyses for patients of other ancestries taking SSRI antidepressants showed mixed results, suggesting a weaker effect within non-Caucasian populations. These results mirror that of Porcelli et al., who found that when analyzing 9 studies of Caucasians taking SSRIs, individuals with the S/S genotype were significantly less likely to respond to SSRI treatment than individuals with the L/L genotype (OR=1.71, p=0.003).5 These findings are similar to the results of a previous meta-analysis, where individuals carrying the S allele were significantly less likely to respond to SSRI treatment.6
The rs25531 SNP has been evaluated in 12 publications to determine its impact on antidepressant efficacy, as described in Table 1.7–18 Four of the 12 studies7,8,12,13, including two independent analyses of STAR*D data8,13, analyzed the rs25531 SNP alone and found no impact of the rs25531 SNP on SSRI efficacy. Eight of the 12 studies assessed the rs25531 SNP in combination with the 5-HTTLPR (i.e. LA vs. LG + S).9–11,14–18 Four of these studies showed that the LA allele was able to predict treatment outcome.9,16–18 However, one of these studies was only significant for anxious depression.9 Additionally, 2 of these 4 studies also analyzed the effect of the 5-HTTLPR alone. They found that analyzing 5-HTTLPR in combination with rs25531 did not add additional specificity to 5-HTTLPR testing alone.9,18 Furthermore, the remaining 4 of 8 studies found no association between the 5-HTTLPR/rs25531 combination and treatment outcome.10,11,14,15
Table 1: Studies Evaluating the Effect of the rs25531 SNP on Antidepressant Efficacy
| Polymorphisms Analyzed | No Association | LA allele able to predict outcome |
| rs25531 | 4 studies (n=3.734)7,8,12,13 | 0 studies |
| 5-HTTLPR/rs25531 | 4 studies (n=350)10,11,14,15 | 4 studies (n=378)9,16-18 |
Is there a connection between SLC6A4 genotype and antidepressant-induced adverse events?
In 2010, Kato et al. performed a meta-analysis of 8 studies (n=2,323) on the 5-HTTLPR and SSRI-induced side effects.19 They found that subjects carrying the S allele (L/S or S/S) had an increased risk of side effects (OR=1.39, p=0.02). However, the impact of the rs25531 SNP on antidepressant-induced side effects is less well studied. In the STAR*D sample (n=1,655), Hu et al.20 found a significant effect of SLC6A4 when using the undifferentiated L/S allele, as well as when differentiating the L allele by the rs25531 SNP (i.e. LA vs. LG + S). Smaller studies examining the impact of the rs25531 SNP have produced mixed results.17,21,22
What is the clinical significance of SLC6A4 genotyping?
The SLC6A4 5-HTTLPR has demonstrated the ability to predict efficacy and adverse events with SSRI treatment in multiple meta-analyses, particularly for patients of Caucasian ancestry. The effect size of the polymorphism (OR=1.71 for efficacy) exceeds the standard set for cost-effectiveness by Perlis et al. (OR=1.5) in a simulation of data from the STAR*D study.23 The 5-HTTLPR polymorphism has also demonstrated cost-effectiveness in two independent cost-effectiveness modeling scenarios.24,25 However, the rs25531 SNP has not demonstrated an impact on antidepressant efficacy, and there is not yet enough data to fully determine its impact on antidepressant-induced adverse events. More data is needed before the rs25531 SNP can be recommended for use in treatment selection.
What treatment options exist for individuals with genetic variation in SLC6A4?
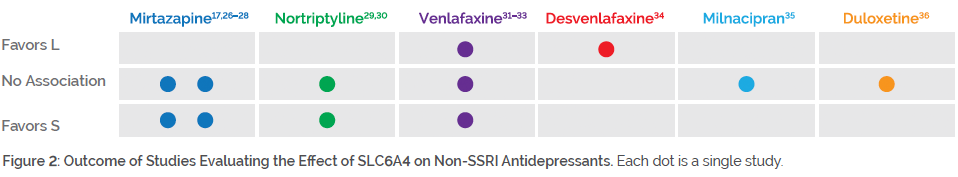
Individuals with genetic variation in SLC6A4 may benefit from medications outside of the SSRI class. As shown in Figure 2, studies on mirtazapine, nortriptyline, venlafaxine, desvenlafaxine, milnacipran, and duloxetine mostly showed no impact of SLC6A4 genotype on efficacy (and in some cases, improved efficacy among S/S individuals).
For example, some studies have focused on mirtazapine, owing to its unique mechanism of action with little affinity for the serotonin transporter. One study found that individuals carrying an S allele had reduced side effects to mirtazapine26, and another study found that those with the S/S genotype responded better to mirtazapine27. Two other studies found no impact of SLC6A4 genotype on mirtazapine response.17,28 For nortriptyline, one study showed that S/S individuals had a favorable response29, while another study failed to find an association30. Studies on venlafaxine have produced mixed results. One study found no impact of SLC6A4 genotype on venlafaxine response31, a second study found poorer response among individuals with the S/S genotype32, and a third study found poorer response among individuals with the LA/LA genotype33.
While these medications are potentially preferential over SSRIs among patients with SLC6A4 variation, they may be influenced by variation in other pharmacokinetic or pharmacodynamic genes. Combinatorial pharmacogenomic testing through GeneSight® Psychotropic analyzes variation at 14 genetic loci (including SLC6A4) and has demonstrated clinical validity, clinical utility, and economic utility in multiple published clinical studies37–42 and three meta-analyses43-45.
References
- Hu, X.-Z. et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am. J. Hum. Genet. 78, 815–826 (2006).
- Martin, J., Cleak, J., Willis-Owen, S. a G., Flint, J. & Shifman, S. Mapping regulatory variants for the serotonin transporter gene based on allelic expression imbalance. Mol. Psychiatry 12, 421–422 (2007).
- Philibert, R. A. et al. Rapid publication: The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa adoption studies. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 147, 543–549 (2008).
- Karlović, D. Serotonin transporter gene (5-HTTLPR) polymorphism and efficacy of selective serotonin reuptake inhibitors–do we have sufficient evidence for clinical practice. Acta Clin. Croat. 52, 353–62 (2013).
- Porcelli, S., Fabbri, C. & Serretti, A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur. Neuropsychopharmacol. 22, 239–58 (2012).
- Serretti, a, Kato, M., De Ronchi, D. & Kinoshita, T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol. Psychiatry 12, 247–57 (2007).
- Kraft, J. B., Slager, S. L., McGrath, P. J. & Hamilton, S. P. Sequence analysis of the serotonin transporter and associations with antidepressant response. Biol. Psychiatry 58, 374–381 (2005).
- Kraft, J. B. et al. Analysis of association between the serotonin transporter and antidepressant response in a large clinical sample. Biol. Psychiatry 61, 734–42 (2007).
- Baffa, A. et al. Norepinephrine and serotonin transporter genes: impact on treatment response in depression. Neuropsychobiology 62, 121–31 (2010).
- Domschke, K. et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int. J. Neuropsychopharmacol. 1–10 (2014). doi:10.1017/S146114571400039X
- Gudayol-Ferré, E. et al. The role of clinical variables, neuropsychological performance and SLC6A4 and COMT gene polymorphisms on the prediction of early response to fluoxetine in major depressive disorder. J. Affect. Disord. 127, 343–351 (2010).
- Shiroma, P. R., Drews, M. S., Geske, J. R. & Mrazek, D. a. SLC6A4 Polymorphisms and Age of Onset in Late-life Depression on Treatment Outcomes with Citalopram: A Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Report. Am. J. Geriatr. Psychiatry 1–9 (2013). doi:10.1016/j.jagp.2013.02.012
- Mrazek, D. a et al. SLC6A4 variation and citalopram response. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 150B, 341–51 (2009).
- Dreimüller, N. et al. The serotonin transporter promoter polymorphism (5-HTTLPR) affects the relation between antidepressant serum concentrations and effectiveness in major depression. Pharmacopsychiatry 45, 108–13 (2012).
- Maron, E. et al. Serotonin transporter promoter region polymorphisms do not influence treatment response to escitalopram in patients with major depression. Eur. Neuropsychopharmacol. 19, 451–6 (2009).
- Gudayol-Ferré, E. et al. Prediction of remission of depression with clinical variables, neuropsychological performance, and serotonergic/dopaminergic gene polymorphisms. Hum. Psychopharmacol. Clin. Exp. 27, 577–586 (2012).
- Staeker, J., Leucht, S., Laika, B. & Steimer, W. Polymorphisms in serotonergic pathways influence the outcome of antidepressant therapy in psychiatric inpatients. Genet. Test. Mol. Biomarkers 18, 20–31 (2014).
- Manoharan, A., Shewade, D. G., Rajkumar, R. P. & Adithan, S. Serotonin transporter gene (SLC6A4) polymorphisms are associated with response to fluoxetine in south Indian major depressive disorder patients. Eur. J. Clin. Pharmacol. 1–6 (2016). doi:10.1007/s00228-016-2099-9
- Kato, M. & Serretti, a. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol. Psychiatry 15, 473–500 (2010).
- Hu, X.-Z. et al. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. Arch. Gen. Psychiatry 64, 783–92 (2007).
- Dombrovski, A. Y. et al. Serotonin transporter triallelic genotype and response to citalopram and risperidone in dementia with behavioral symptoms. Int. Clin. Psychopharmacol. 25, 37–45 (2010).
- Garfield, L. D. et al. Common Selective Serotonin Reuptake Inhibitor Side Effects in Older Adults Associated with Genetic Polymorphisms in the Serotonin Transporter and Receptors: Data from a Randomized Controlled Trial. Am. J. Geriatr. Psychiatry 1–9 (2013). doi:10.1016/j.jagp.2013.07.003
- Perlis, R. H., Patrick, A., Smoller, J. W. & Wang, S. When is Pharmacogenetic Testing for Antidepressant Response Ready for the Clinic? A Cost-effectiveness Analysis Based on Data from the STAR*D Study. Neuropsychopharmacology 34, 2227–2236 (2012).
- Olgiati, P., Bajo, E., Bigelli, M., De Ronchi, D. & Serretti, A. Should pharmacogenetics be incorporated in major depression treatment? Economic evaluation in high- and middle-income European countries. Prog. Neuropsychopharmacol. Biol. Psychiatry 36, 147–54 (2012).
- Serretti, A., Olgiati, P., Bajo, E., Bigelli, M. & De Ronchi, D. A model to incorporate genetic testing (5-HTTLPR) in pharmacological treatment of major depressive disorders. World J. Biol. Psychiatry 12, 501–15 (2011).
- Murphy, G. M., Hollander, S. B., Rodrigues, H. E., Kremer, C. & Schatzberg, A. F. Effects of the serotonin transporter gene promoter polymorphism on mirtazapine and paroxetine efficacy and adverse events in geriatric major depression. Arch. Gen. Psychiatry 61, 1163–9 (2004).
- Kang, R.-H., Wong, M.-L., Choi, M.-J., Paik, J.-W. & Lee, M.-S. Association study of the serotonin transporter promoter polymorphism and mirtazapine antidepressant response in major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 31, 1317–1321 (2007).
- Chang, H. S. et al. Interaction of 5-HTT and HTR1A gene polymorphisms in treatment responses to mirtazapine in patients with major depressive disorder. J. Clin. Psychopharmacol. 34, 446–54 (2014).
- Kim, H. et al. Monoamine transporter gene polymorphisms and antidepressant response in koreans with late-life depression. JAMA 296, 1609–18 (2006).
- Pollock, B. G. et al. Allelic variation in the serotonin transporter promoter affects onset of paroxetine treatment response in late-life depression. Neuropsychopharmacology 23, 587–90 (2000).
- Ng, C. et al. Pharmacogenetic polymorphisms and response to escitalopram and venlafaxine over 8 weeks in major depression. Hum. Psychopharmacol. 28, 516–22 (2013).
- Lee, S.-H. et al. Serotonin transporter gene polymorphism associated with short-term treatment response to venlafaxine. Neuropsychobiology 62, 198–206 (2010).
- Proft, F. et al. SLC6A2 and SLC6A4 Variants interact with Venlafaxine Serum Concentrations to Influence Therapy Outcome. Pharmacopsychiatry 47, 245–50 (2014).
- Ng, C. H., Bousman, C., Smith, D. J. & Dowling, N. A Prospective Study of Serotonin and Norepinephrine Transporter Genes and the Response to Desvenlafaxine Over 8 Weeks in Major Depressive Disorder. 2–4 (2016).
- Yoshida, K. et al. Prediction of antidepressant response to milnacipran by norepinephrine transporter gene polymorphisms. Am. J. Psychiatry 161, 1575–1580 (2004).
- Perlis, R. H., Fijal, B., Dharia, S., Heinloth, A. N. & Houston, J. P. Failure to replicate genetic associations with antidepressant treatment response in duloxetine-treated patients. Biol. Psychiatry 67, 1110–3 (2010).
- Hall-Flavin, D. K. et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl. Psychiatry 2, e172 (2012).
- Hall-Flavin, D. K. et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet. Genomics 23, 535–48 (2013).
- Winner, J. G., Carhart, J. M., Altar, C. A., Allen, J. D. & Dechairo, B. M. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov. Med. 16, 219–27 (2013).
- Greden, J.F. et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: A large, patient- and rater-blinded, randomized, controlled study. J. Psychiatr. Res. 111, 59-67 (2019).
- Winner, J., Allen, J. D., Anthony Altar, C. & Spahic-Mihajlovic, a. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl. Psychiatry 3, e242 (2013).
- Winner, J. G. et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a one year prospective evaluation. Curr. Med. Res. Opin. 1–30 (2015). doi:10.1185/03007995.2015.1063483
- Altar, C. A. et al. Clinical Utility of Combinatorial Pharmacogenomics-Guided Antidepressant Therapy: Evidence from Three Clinical Studies. Mol. Neuropsychiatry 1–11 (2015). doi:10.1159/000430915
- Altar, C. A. et al. Clinical validity : Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. 1–9 (2015). doi:10.1038/tpj.2014.85
- Brown, L. et al. The clinical utility of combinatorial pharmacogenomic testing for patients with depression: a meta-analysis. Pharmacogenomics 21(8):559-569 (2020).