Understanding the GeneSight test algorithm
The GeneSight test uses a unique combinatorial algorithm, which evaluates how variations in multiple genes may influence an individual’s outcomes with certain medications.
The algorithm provides important information regarding how an individual may metabolize and/or respond to certain medications commonly prescribed to treat depression, anxiety, ADHD and other psychiatric conditions. This information supplements a healthcare provider’s comprehensive medical evaluation to help personalize treatment plans.
The GeneSight Psychotropic test has been evaluated in multiple published, peer-reviewed clinical studies. It is the only neuropsychiatric pharmacogenomic test backed by such extensive research.
The role of genetics in predicting outcomes with medications used for psychiatric conditions
Understanding the combinatorial approach of the GeneSight test starts with understanding our basic genetics: Each individual has a unique collection of genes responsible for their various genetic traits.
The GeneSight test analyzes genetic variation found in 14 total genes that may impact a patient’s outcomes with certain medications. This includes 9 pharmacokinetic genes and 5 pharmacodynamic genes:
- Pharmacokinetic (PK) genes are involved in how the body metabolizes, or breaks down, a particular medication through specific drug metabolizing enzymes.
- Pharmacodynamic (PD) genes provide information on how your DNA may impact your likelihood of response or risk for side effects with some medications.
The GeneSight test takes into consideration all clinically relevant genes known to affect metabolism or response to a medication and assigns weights based on how big (or small) of a role it is expected to play in the medication’s overall metabolism or mechanism of action.
The data that informs the algorithm
Evidence used to build the algorithm comes from comprehensive literature and data reviews. Sources include published scientific literature (collected from PubMed, Embase, Google Scholar, etc.), approved regulatory drug labels, and review applications (FDA). Animal-derived, preclinical data is not included – only human-based data.
The data can be separated into three categories:
- FDA-reviewed pharmacokinetic (PK) and pharmacodynamic (PD) documents submitted as part of a new drug application (NDA).
- in-vitro (test tube) and in-vivo (human) data assessing medication pathways that have been published in peer-reviewed journals. This identifies the genes involved in the metabolism and/or the mechanism of action of a medication.
- in-vitro and in-vivo data evaluating genetic variations and applying expected activity levels (i.e., phenotype expression based on genetic variation) that have either been published in peer-reviewed journals or derived from internal lab data. This identifies to what extent the genetic variation is expected to impact the metabolism and/or the mechanism of action of a medication.
The quality of the data is scrutinized, with higher quality evidence having a greater impact in the algorithm. For example, in-vivo data is held in higher regard than in-vitro data.
Additionally, data assessed from evidence-grading bodies such as the Pharmacogenomics Knowledgebase (PharmGKB), the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetic Working Group (DPWG) are also evaluated for inclusion. This ensures that all sources of clinically actionable gene-drug interactions (GDI) and genotype-phenotype relationships have been considered. It is important to note that our evaluation of the data may lead to different interpretations than what is concluded by individual expert PGx groups, evidence grading bodies, or other PGx companies.
The field of pharmacogenomics is constantly evolving. As the evidence surrounding the impact of established and newly identified genetic variants on medication metabolism and response evolves, so too will the algorithm. The Myriad team continually reviews available data to determine if and how it impacts the GeneSight proprietary algorithm. A full review of the GeneSight test is completed approximately every 18 months to 2 years.
The GeneSight algorithm
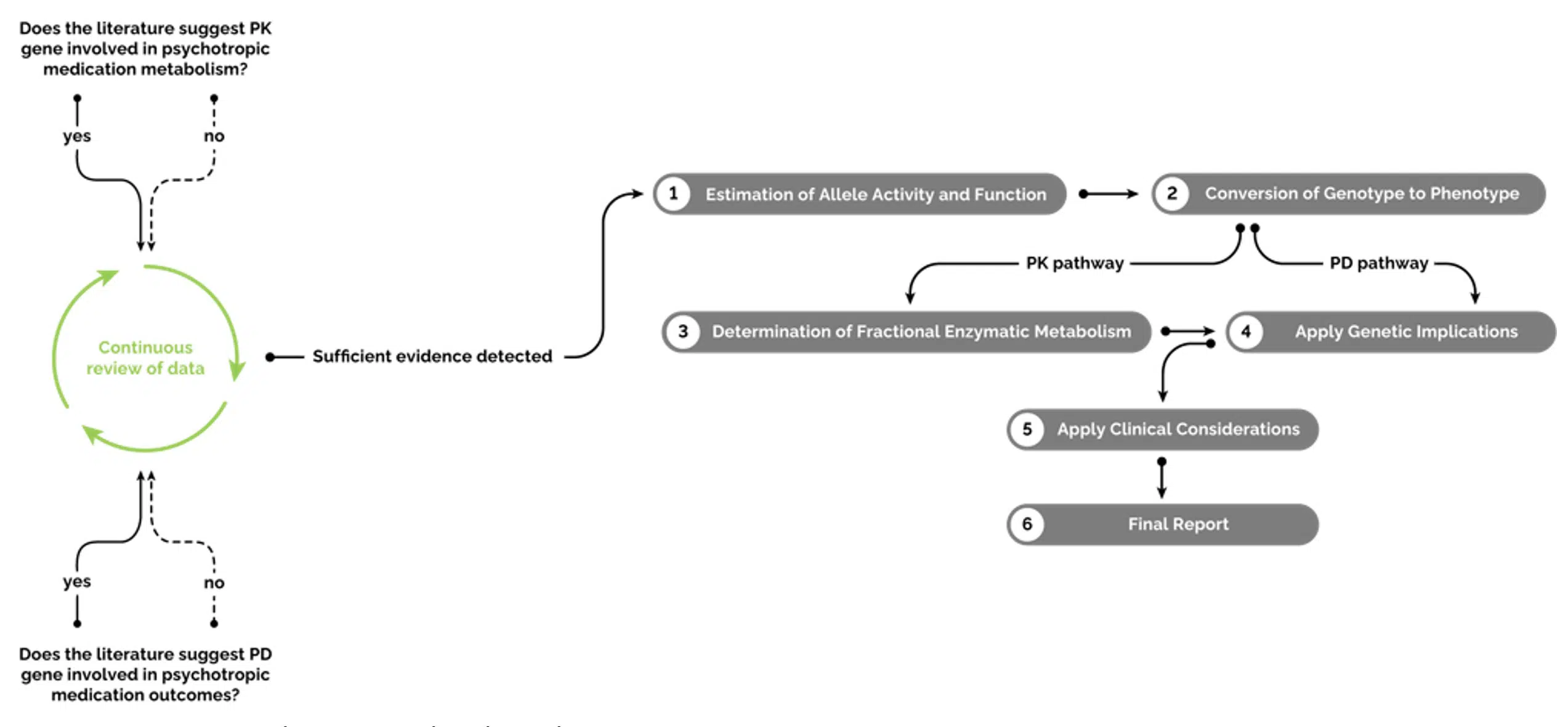
Figure 1: The GeneSight Algorithm Process
1: Estimation of allele activity and function
An allele refers to different versions of a gene. Each allele that makes up a genotype is assigned an expected activity level or function.
For pharmacokinetic genes, *1 refers to the normal or functional allele. A different activity level, such as increased, decreased, or no activity may be assigned if there is a sufficient body of evidence in published literature.
For pharmacodynamic genes, an allele may be associated with a different function, such as increased risk of side effects or reduced response, if there are several consistent and well-replicated findings.
2: Conversion of genotype to phenotype
An individual’s combination of alleles for a specific gene is the genotype. The effect of that specific combination is the phenotype.
For pharmacokinetic genes, the phenotype determines the metabolizer status, based on the detected alleles and expected total percent enzyme activity, which is calculated by adding together the percent enzyme activity of each individual allele.
On the GeneSight test, the metabolizer status can be one of four phenotype groups (i.e., activity level groups):
- Poor Metabolizer: Medication is broken down very slowly. May experience side effects at standard doses.
- Intermediate Metabolizer: Slow rate of metabolism. May have too much medication at standard doses, potentially causing side effects.
- Extensive Metabolizer: Normal rate of metabolism. Has normal amount of medication at standard doses.
- Ultrarapid Metabolizer: Medication is rapidly broken down. Medication may be removed from a patient’s system too quickly to provide symptom relief.
For pharmacodynamic genes, the phenotype would depict the likelihood of response or the risk of side effects and is dependent on which alleles are detected. On the GeneSight test, the likelihood of response would either be considered normal or reduced, and the risk of side effects would either be considered the standard risk or an elevated risk, depending on the respective PD gene and the genotype expressed.
3: Determination of percentage of enzyme involvement
The next step is to determine the percentage of enzyme involvement in the metabolism of a medication, as the majority of medications are broken down via multiple enzymatic pathways. Pharmacokinetic genes contain the information needed to make these metabolic enzymes. Therefore, this step only applies to pharmacokinetic genes.
How much an enzyme is expected to affect an individual medication’s metabolism is approximated based on the literature. The assigned percentages (i.e., enzyme weight) are proprietary to the GeneSight algorithm. However, the enzymes that are involved in the metabolism of each medication are depicted at the end of every GeneSight report in the Gene-Drug Interaction (GDI) chart for informational purposes.
Once the weighted pathways are determined, an individual’s phenotype for each relevant gene can be applied to the algorithm, which will provide information on how variations in pharmacokinetic enzymes may affect medication metabolism.
4: Application of genetic implications
To convey all potential outcomes and all possibilities, the algorithm is run using the relevant weights from the pathways involved and the individual’s phenotypes. Both pharmacodynamic and pharmacokinetic genes play important roles in outcomes with medications. However, not all medications will be affected by both kinds of genes.
Based on a patient’s genetic variation, medications can be placed into the green “Use as Directed” category, the yellow “Moderate Gene-Drug Interaction” category or the red “Significant Gene-Drug Interaction” category. The following example of Drug X conceptually depicts this step (Figure 3).
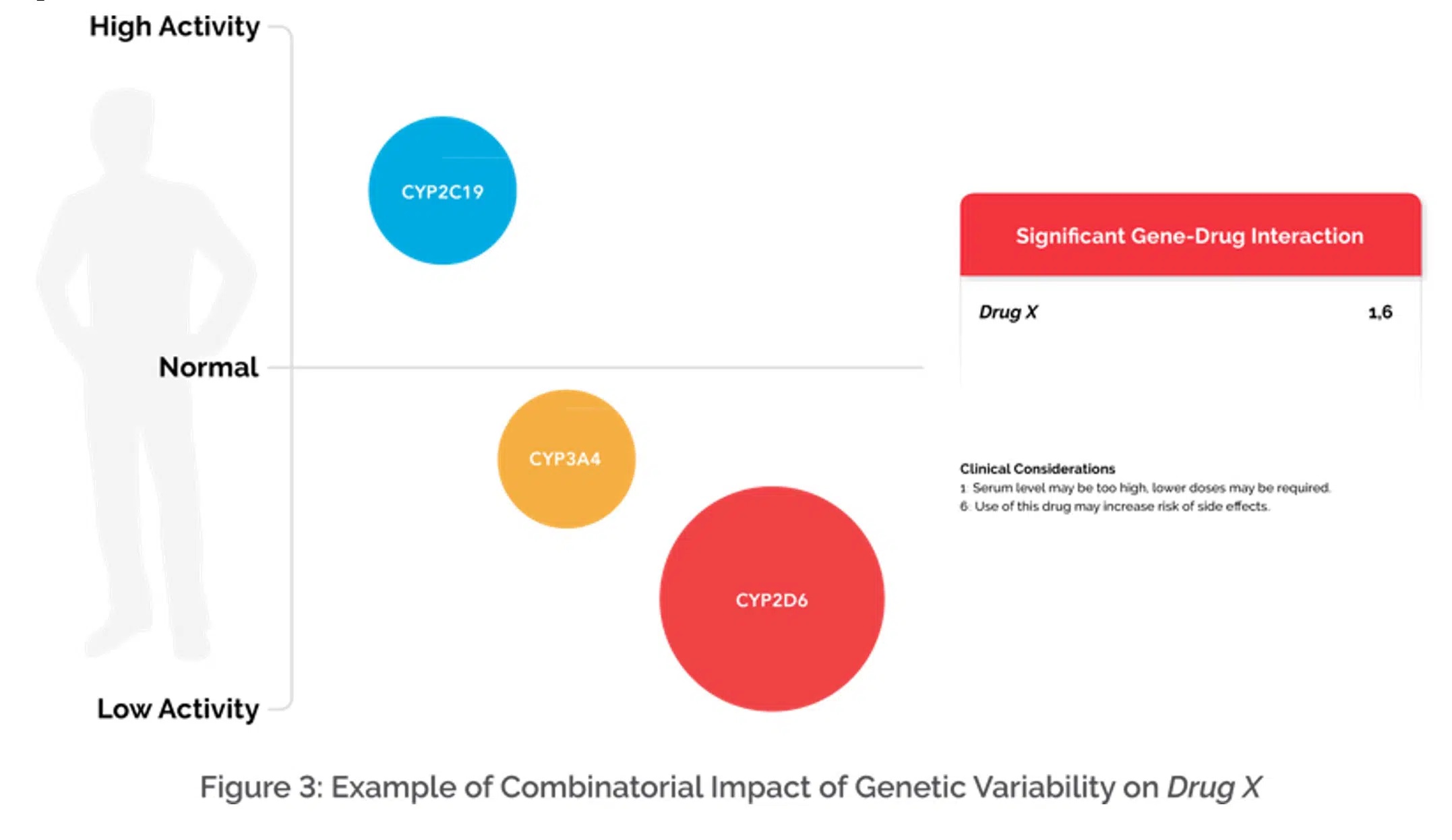
Figure 3: Illustrates an example that shows multiple genes involved in the metabolism of Drug X and the size of the circle indicates the relative weight of the gene. Based on the combination of genes, the relative weight of each gene, and genetic variation detected, the algorithm is then able to categorize a medication on the GeneSight report and apply clinical considerations when applicable.
5: Application of clinical considerations
If a medication does have moderate or significant gene-drug interactions (GDIs), then specific clinical considerations are added to the report for the applicable medications (Figure 4).
Clinical considerations share the rationale for a medication’s classification. They can help inform a clinician’s treatment decisions by providing information about how an individual’s genes may impact outcomes with certain medications.

Figure 4: Examples of some of the clinical considerations on the GeneSight Psychotropic report.
6: Generation of final GeneSight report
The final step is to generate the GeneSight test results report. The final report includes all 64 medications placed into one of three categories, which include:
- Green – “Use as Directed”
- Yellow – “Moderate Gene-Drug Interaction”
- Red – “Significant Gene-Drug Interaction”
While the color coding provides easy-to-digest information, there’s more to the report than just these categories. Medication failure may still occur with medications in the “Use as Directed” category, and medications in the “Significant GDI” category may be effective, as all medications have unique efficacy and side effect profiles.
“We often use a stoplight analogy when talking with healthcare providers and patients,” says Holly Johnson, Ph.D., medical information director, Myriad Neuroscience. She explains:
“Think of an intersection with a stoplight as the point of medication decision making. You can go in any number of directions.
If the light is green, you can pass through the intersection as you would normally drive. Likewise, with the GeneSight test, when medications fall in the green ‘Use as Directed’ category, it means that there are no genetic variations in the tested genes that are expected to impact the patient’s outcomes with those medications. However, like at a green light, there can be other factors that could cause problems such as other cars or debris on the road. Similarly, there can be other factors that may impact your ability to metabolize or respond to the medications listed in the green category, such as drug/drug interactions, food/drug interactions, allergies and other factors.
The yellow ‘Moderate Gene-Drug Interaction’ category is like a yellow light – the healthcare provider may want to slow down and be more cautious about how that medication is used. Dosage adjustments, closer monitoring, or other strategies may be necessary.
Red category medications, or those in the ‘Significant Gene-Drug Interaction’ category, can be treated just like a red light: You should stop, but it doesn’t mean you can’t go through the intersection (e.g., sometimes it is OK to turn right on red). However, the healthcare provider needs to carefully consider the information that is being presented by the clinical considerations.”
The GeneSight test can be one part of a doctor’s evaluation
Pharmacogenomic testing, like the GeneSight test, is intended to provide insight on how an individual’s genetic variation may affect outcomes with certain medications and should be used in conjunction with other clinical factors, expertise, and judgment. Therefore, pharmacogenomic tests like GeneSight are intended to be ordered by and used only in consultation with a healthcare provider who can prescribe medications.
The GeneSight test results are designed to be just one part of a larger, complete patient assessment, which would include proper diagnosis and consideration of an individual’s medical history, other medications they may be taking, their family history, and other factors.
Because the results in the final report are specific to the genetic profile of the individual patient and provide insight into how an individual may metabolize or respond to specific medications that are used to treat mental health conditions like depression, the GeneSight Psychotropic test and its algorithm can be a valuable tool in the healthcare provider’s toolbox.

