In multiple previous studies, the GeneSight test, a combinatorial multigene pharmacogenomic test, powered by CPGx technology, has shown an ability to predict poorer antidepressant outcomes and to help guide healthcare providers to more genetically optimal medications, leading to improved patient outcomes.1–5
What is “combinatorial pharmacogenomics”?
Neuropsychiatric medications are metabolized by multiple enzymes and interact with multiple neuropathways in order to attain clinical response.6 Pharmacogenomic testing has traditionally made a necessary, but flawed, pharmacologic assumption — that only a single drug-metabolizing enzyme is relevant for a given medication. This assumption was likely made because the vast number of permutations of genetic effects and metabolic pathways rendered the interpretation of test results unfeasible to implement into clinical practice. The GeneSight test’s proprietary CPGx approach was specifically designed to address this problem. Results from over 750 published clinical studies, detailed pharmacology, manufacturers’ FDA-approved medication labels and our proprietary clinical research have been integrated to analyze and weight the importance of multiple pharmacokinetic and pharmacodynamic genes. The factors taken into consideration include:
- All known CYP450 and non-CYP450 metabolic pathways of each medication and medication metabolite, weighted for their relative importance
- The pharmacodynamic activity levels of the parent compound and any active metabolites and genetic variation that may impact pharmacodynamic activity, incorporating known clinical considerations (e.g. hydroxybupropion’s seizure risk mentioned below)
- Validated research regarding all known functionally significant alleles in all relevant weighted genes
- FDA labeling information related to genetically mediated efficacy or tolerability of a medication
The GeneSight test then integrates this information with the genetic test results obtained from each patient’s buccal swab in order to categorize the 64 FDA-approved medications into the three “traffic light” color-coded categories in our GeneSight report revealing ascending levels of gene-drug interactions: green (use as directed), yellow (moderate gene-drug interaction) and red (significant gene-drug interaction). This integrated approach accounts for multiple pharmacokinetic and pharmacodynamic pathways and categorizes them according to severity of their gene-drug interaction as a whole construct, as opposed to a reductionistic “one gene, one drug” approach.
Do you have some examples of the clinical significance of combinatorial pharmacogenomic testing?
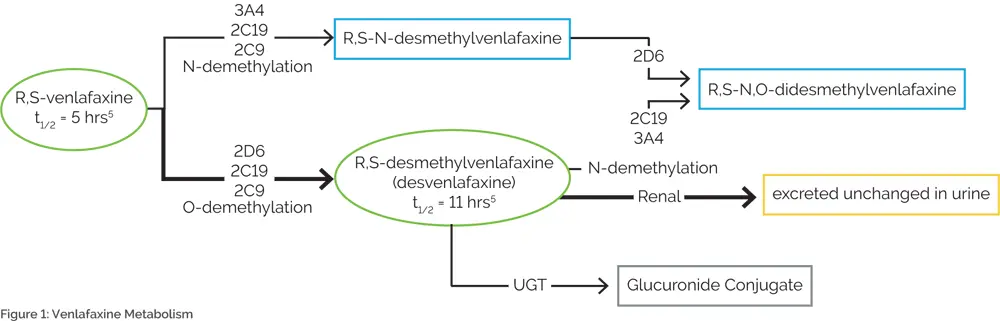
Venlafaxine is an excellent example of the importance of combinatorial pharmacogenomic testing. While several sources cite CYP2D6 as the only relevant CYP450 enzyme7,8, abundant data have demonstrated the clinical relevance of other enzymes, including CYP2C19, CYP2C9, and CYP3A49–13 (Figure 1). Multiple case studies have reported on venlafaxine-induced adverse events among patients with reduced activity at multiple CYP450 enzymes.14–17 One of these cases involved an individual who was a CYP2D6 extensive metabolizer, but was a CYP2C19 intermediate metabolizer and taking a medication that inhibited CYP2C9. The deficiencies at these two pathways resulted in severe tremor, which abated when venlafaxine was discontinued.17 Another case describes a poor metabolizer of both CYP2D6 and CYP2C19 who died of venlafaxine poisoning following a 150 mg dose of extended-release venlafaxine.16
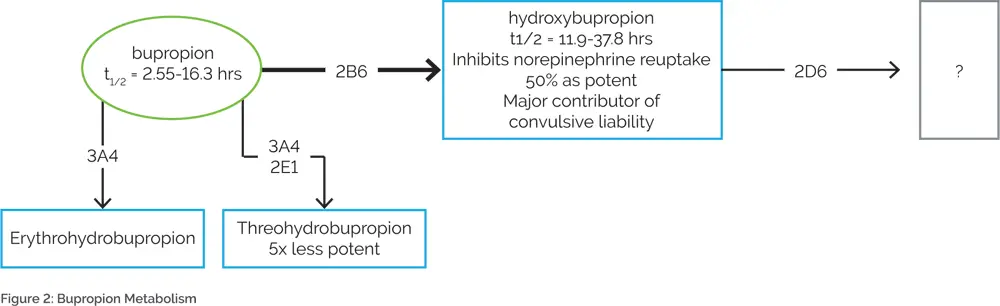
Another example of the importance of combinatorial pharmacogenomic testing is bupropion. Bupropion is metabolized by CYP2B6 to hydroxybupropion18–20, which is further metabolized by CYP2D621 (Figure 2). Hydroxybupropion is thought to be the major contributor to the seizure risk associated with bupropion.22,23 Combinatorial pharmacogenomic testing would predict that individuals who are combined CYP2B6 ultrarapid metabolizers and CYP2D6 poor metabolizers may be expected to have higher hydroxybupropion levels, leading to an increased risk of bupropion-induced seizures. Conversely, traditional single-gene testing for only CYP2B6 may predict that a CYP2B6 ultrarapid metabolizer would need a higher dose of bupropion. However, knowledge of CYP2D6 genotype is also necessary since a patient may be at an increased risk of seizures if hydroxybupropion is metabolized slowly. Therefore, increasing the dose of bupropion in this case would be ill-advised.
Does combinatorial pharmacogenomics produce better patient outcomes than single-gene testing?
Single-gene pharmacogenomic testing has historically produced mixed results regarding clinical outcomes24, although recent guidelines have brought attention to clinically important single gene-drug interactions25-28. One viable reason for the lack of overwhelming evidence for single-gene testing is that the combined (i.e. combinatorial) effects of multiple genes give the most accurate pharmacologic picture for a given medication. Without this full combinatorial picture, the clinical effect may be muted creating mixed outcomes. An important clinical question is, “Do the additive effects of the combinatorial process predict improved patient outcomes compared to each gene individually?”
To address this question, Assurex Health published an analysis comparing the predictive ability of the GeneSight test to traditional single-gene testing in 201529. The analysis used combined data from the standard of care arms of three prospective clinical outcome trials of the GeneSight test. It is important to note that this analysis assessed the standard of care (i.e. control arm or treatment as usual) group of patients so that no patient or patient’s healthcare provider was aware of their pharmacogenomic information, thus creating blinded outcomes.
In this analysis, patients were stratified into subgroups based on their medication regimen, accounting for each medication’s combined metabolic pathway. For example, if patient 1 was taking venlafaxine, which is a substrate for CYP2D6, CYP2C19 and CYP2C9, she was included in the CYP2D6, CYP2C19, and CYP2C9 analyses. If patient 2 was taking venlafaxine and also taking olanzapine, which is metabolized through CYP1A2 and CYP2D6, he would be included in the CYP2D6, CYP2C19, CYP1A2, and CYP2C9 analyses.
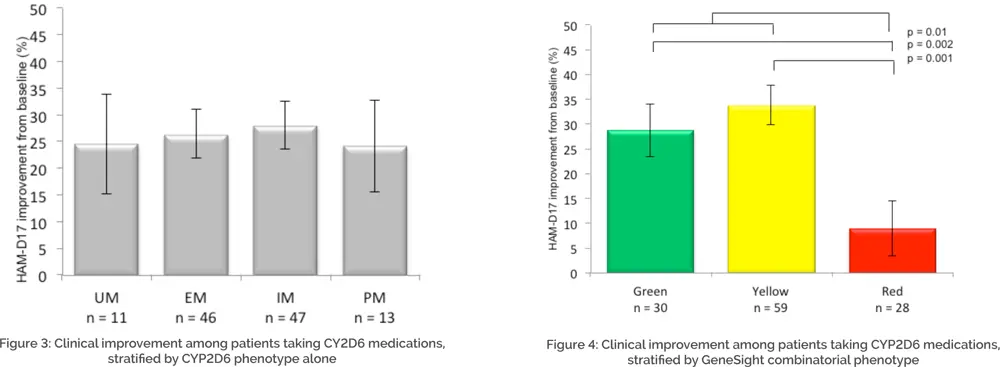
Once these subgroups were assigned, traditional single-gene phenotypes (e.g. CYP2D6 UM, EM, IM, and PM) were assessed for improvement in clinical outcomes. The results of the CYP2D6 subgroup analysis are displayed in Figure 3. When only CYP2D6 genotype was considered for medications that were primarily (though not exclusively) metabolized by CYP2D6, the resultant phenotypes were not predictive of patient outcomes, suggesting that individual gene testing alone is unable to accurately predict patient response to medications, agreeing with the history of outcome data for these tests.24
The same analysis was performed using the phenotype categories from the GeneSight test’s proprietary CPGx® process (i.e. “green,” “yellow,” “red”) instead of each phenotype (UM, EM, IM, and PM). The categorical GeneSight process (green, yellow, red) reveals combined information about all relevant genes so that more information may be clinically assessed than simply the ‘primary metabolic pathway’ for a given medication. The results of the GeneSight test’s proprietary CPGx analysis are displayed in Figure 4. When using the GeneSight combinatorial report to predict response to medications that were primarily (though not exclusively) metabolized by CYP2D6, significant differences were observed, with the red category showing lower improvement in depressive symptoms than subjects in the green or yellow categories. Given that these patients were fully blinded to their pharmacogenomic information, this demonstrates the ability of the GeneSight test to a priori predict patient medication response when all of the pharmacogenomic information is integrated and presented in an actionable form. Without knowing which medications a patient will do poorly on, a healthcare provider cannot do something different for the patient. The GeneSight test helps guide healthcare providers to select more genetically optimal medications for their patients.
Conclusion
The GeneSight test employs a proprietary and wholly unique method of predicting patient medication response using a combinatorial pharmacogenomic method. Combinatorial pharmacogenomics uses knowledge of each medication’s unique set of pharmacokinetic and pharmacodynamic characteristics to incorporate and appropriately weight genetic variation at multiple loci to produce more accurate predictions of patient response than testing solely for the primary metabolic pathway of a medication.
References
- Hall-Flavin, D. K. et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl. Psychiatry 2, e172 (2012).
- Hall-Flavin, D. K. et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet. Genomics 23, 535–48 (2013).
- Winner, J., Allen, J. D., Anthony Altar, C. & Spahic-Mihajlovic, a. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl. Psychiatry 3, e242 (2013).
- Winner, J. G., Carhart, J. M., Altar, C. A., Allen, J. D. & Dechairo, B. M. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov. Med. 16, 219–27 (2013).
- Winner, J. G. et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a one year prospective evaluation. Curr. Med. Res. Opin. 1–30 (2015). doi:10.1185/03007995.2015.1063483
- Mrazek, D. A. Psychiatric Pharmacogenomics. (Oxford University Press, 2010).
- Wyeth Pharmaceuticals. Venlafaxine Prescribing Information. (2012). at
- Cerner Multum Inc. Venlafaxine Drug Monograph. (2014). at
- McAlpine, D. E. et al. Effect of cytochrome P450 enzyme polymorphisms on pharmacokinetics of venlafaxine. Ther. Drug Monit. 33, 14–20 (2011).
- Ereshefsky, L. & Dugan, D. Review of the pharmacokinetics, pharmacogenetics, and drug interaction potential of antidepressants: focus on venlafaxine. Depress. Anxiety 12 Suppl 1, 30–44 (2000).
- Fogelman, S. M. et al. O- and N-demethylation of venlafaxine in vitro by human liver microsomes and by microsomes from cDNA-transfected cells: effect of metabolic inhibitors and SSRI antidepressants. Neuropsychopharmacology 20, 480–90 (1999).
- Kirchheiner, J. et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol. Psychiatry 9, 442–73 (2004).
- Fukuda, T. et al. The impact of the CYP2D6 and CYP2C19 genotypes on venlafaxine pharmacokinetics in a Japanese population. Eur. J. Clin. Pharmacol. 56, 175–80 (2000).
- Chua, E. W., Foulds, J., Miller, A. L. & Kennedy, M. a. Novel CYP2D6 and CYP2C19 variants identified in a patient with adverse reactions towards venlafaxine monotherapy and dual therapy with nortriptyline and fluoxetine. Pharmacogenet. Genomics 23, 494–7 (2013).
- Vinetti, M. et al. Severe acute cardiomyopathy associated with venlafaxine overdose and possible role of CYP2D6 and CYP2C19 polymorphisms. Clin. Toxicol. 49, 865–869 (2011).
- Jornil, J. et al. A poor metabolizer of both CYP2C19 and CYP2D6 identified by mechanistic pharmacokinetic simulation in a fatal drug poisoning case involving venlafaxine. Forensic Sci. Int. 226, e26–e31 (2013).
- Geber, C., Ostad Haji, E., Schlicht, K., Hiemke, C. & Tadić, A. Severe tremor after cotrimoxazole-induced elevation of venlafaxine serum concentrations in a patient with major depressive disorder. Ther. Drug Monit. 35, 279–82 (2013).
- Kirchheiner, J. et al. Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13, 619–26 (2003).
- Zhu, a Z. X. et al. CYP2B6 and bupropion’s smoking-cessation pharmacology: the role of hydroxybupropion. Clin. Pharmacol. Ther. 92, 771–7 (2012).
- Benowitz, N. L., Zhu, A. Z. X., Tyndale, R. F., Dempsey, D. & Jacob, P. Influence of CYP2B6 genetic variants on plasma and urine concentrations of bupropion and metabolites at steady state. Pharmacogenet. Genomics 23, 135–41 (2013).
- Pollock, B. G., Sweet, R. A., Kirshner, M. & Reynolds, C. F. Bupropion plasma levels and CYP2D6 phenotype. Ther. Drug Monit. 18, 581–5 (1996).
- Friel, P. N., Logan, B. K. & Fligner, C. L. Three fatal drug overdoses involving bupropion. J. Anal. Toxicol. 17, 436–438 (1993).
- Silverstone, P. H., Williams, R., McMahon, L., Fleming, R. & Fogarty, S. Convulsive liability of bupropion hydrochloride metabolites in Swiss albino mice. Ann. Gen. Psychiatry 7, 19 (2008).
- Matchar, D. B. et al. Testing for cytochrome P450 polymorphisms in adults with non-psychotic depression treated with selective serotonin reuptake inhibitors (SSRIs). Evid. Rep. Technol. Assess. (Full. Rep). 9, 1–77 (2007).
- Hicks, J. K. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. n/a–n/a (2015). at
- Hicks, J. K. et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants. Clin. Pharmacol. Ther. 93, 402–8 (2013).
- Swen, J. J. et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011).
- Conrado, D. J., Rogers, H. L., Zineh, I. & Pacanowski, M. a. Consistency of drug-drug and gene-drug interaction information in US FDA-approved drug labels. Pharmacogenomics 14, 215–23 (2013).
- Altar, C. A. et al. Clinical validity : Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. 1–9 (2015). doi:10.1038/tpj.2014.85